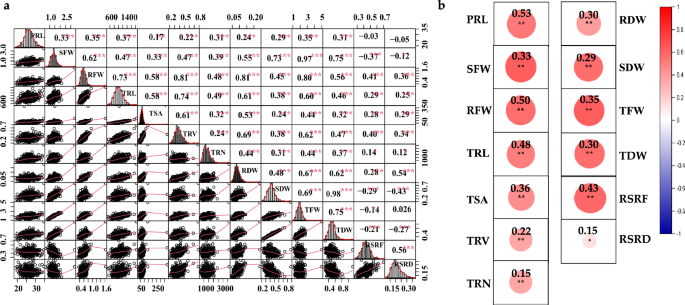
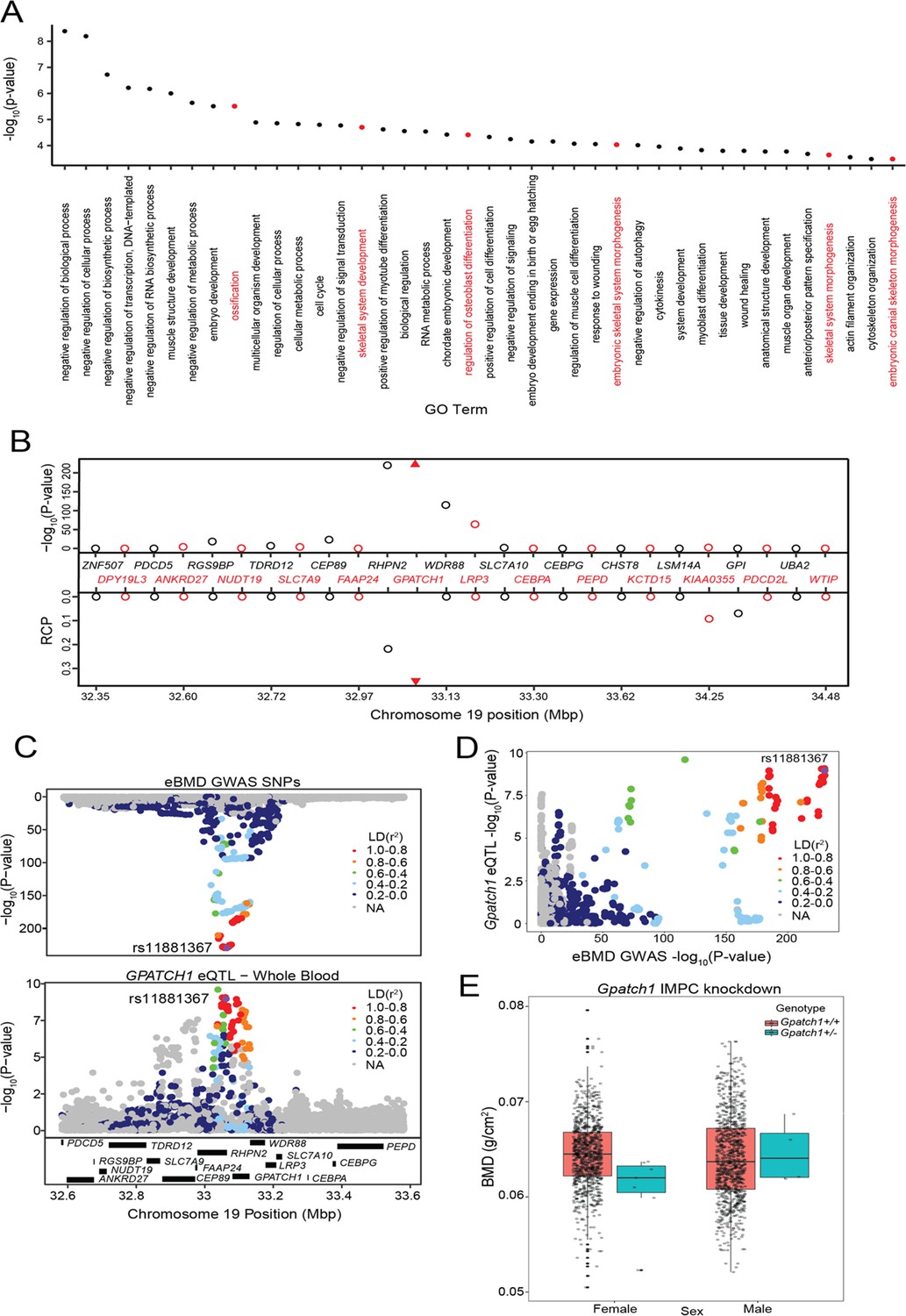
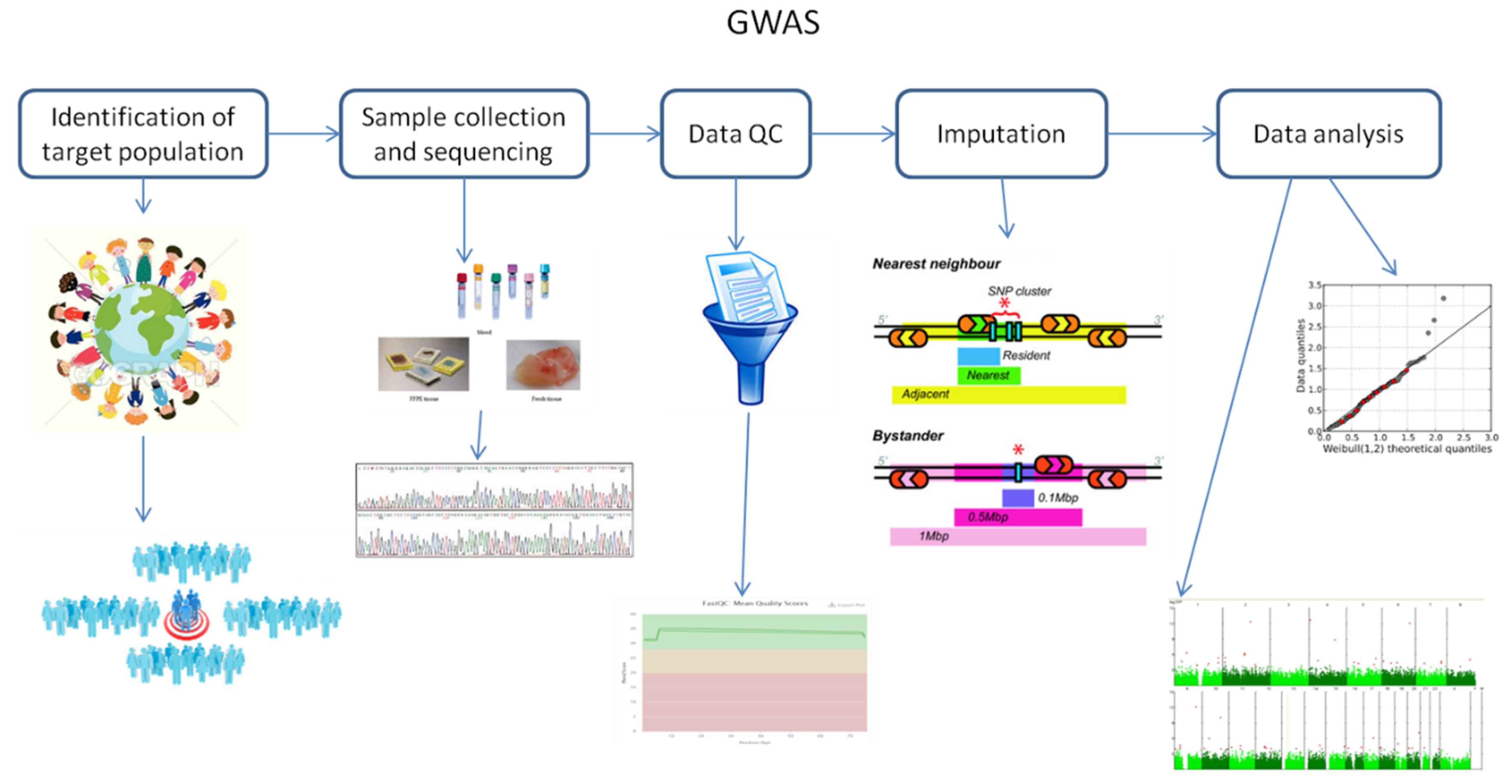
Background Rapeseed (Brassica napus L.) is an essential source of edible oil and livestock feed, as well as a promising source of biofuel. Breeding crops with an ideal root system architecture (RSA) for high phosphorus use efficiency (PUE) is an effective way to reduce the use of phosphate fertilizers. However, the genetic mechanisms that underpin PUE in rapeseed remain elusive. To address this, we conducted a genome-wide association study (GWAS) in 327 rapeseed accessions to elucidate the genetic variability of 13 root and biomass traits under low phosphorus (LP; 0.01 mM P +). Furthermore, RNA-sequencing was performed in root among high/low phosphorus efficient groups (HP1/LP1) and high/low phosphorus stress tolerance groups (HP2/LP2) at two-time points under control and P-stress conditions. Results Significant variations were observed in all measured traits, with heritabilities ranging from 0.47 to 0.72, and significant correlations were found between most of the traits. There were 39 significant trait–SNP associations and 31 suggestive associations, which integrated into 11 valid quantitative trait loci (QTL) clusters, explaining 4.24–24.43% of the phenotypic variance observed. In total, RNA-seq identified 692, 1076, 648, and 934 differentially expressed genes (DEGs) specific to HP1/LP1 and HP2/LP2 under P-stress and control conditions, respectively, while 761 and 860 DEGs common for HP1/LP1 and HP2/LP2 under both conditions. An integrated approach of GWAS, weighted co-expression network, and differential expression analysis identified 12 genes associated with root growth and development under LP stress. In this study, six genes (BnaA04g23490D, BnaA09g08440D, BnaA09g04320D, BnaA09g04350D, BnaA09g04930D, BnaA09g09290D) that showed differential expression were identified as promising candidate genes for the target traits. Conclusion 11 QTL clusters and 12 candidate genes associated with root and development under LP stress were identified in this study. Our study's phenotypic and genetic information may be exploited for genetic improvement of root traits to increase PUE in rapeseed.
Transcriptome-wide association study and eQTL colocalization

Turning genome-wide association study findings into opportunities

Integrating transcription factor occupancy with transcriptome-wide

Unlocking the secrets of our genes: Best practices in genome-wide association studies

Candidate gene association analysis of GRMZM2G075104. a Significant
IJMS, Free Full-Text

Full article: Genome- and transcriptome-wide association studies

QTL mapping of seedling biomass and root traits under different nitrogen conditions in bread wheat (Triticum aestivum L.)

After a decade of genome-wide association studies, a new phase of

Summarizing mapped reads into a gene level count. (a) Mapped reads from

QTL mapping of seedling biomass and root traits under different nitrogen conditions in bread wheat (Triticum aestivum L.)

Integrating single-cell sequencing data with GWAS summary







